What we Do:
Using cutting-edge technology to unlock the secrets of immunity and address global health issues.
Our Approach
Studying immunity in humans promises insights into disease mechanisms with direct translational impact. Previous attempts to explore immune responses in humans were hampered by small sample volumes, diversity between individuals, and the immune system’s complexity.
Technological advances helped overcome many of these obstacles, promoting the field of human systems biology. Systems biology leverages high-throughput techniques (e.g., scRNA-seq, ATAC-seq, CyTOF, Olink) to simultaneously measure many different molecules and cell types in a single blood sample.
Researchers use computational and machine learning tools to analyze this data and assemble a comprehensive model of the unfolding immune response. These models are then queried to identify interactions between various immune system components, to determine disturbed pathways during disease, and to propose mechanistic hypotheses.
In a final step, generated hypotheses are validated using complex human organoid systems or human clinical trials.
We and others have used systems biology approaches to determine vaccine efficacy and durability and to identify mechanisms of immunity to infections, such as COVID-19, and vaccines.
We answer questions in fundamental and translational immunology using systems biology tools. Our goal is to understand how the immune system fights cancer and infections and what happens when it is forced to battle both at the same time.
Our Goals
1
Deciphering Unique Immune Landscapes
2
Crafting Vaccines for Vulnerable Demographics
3
Setting New Standards in Immunological Research
Our Philosophy:
In our lab, we nurture future leaders in immunology, emphasizing creativity, independence, and collaborative excellence. With diverse expertise and a global network, we are collectively driving forward crucial discoveries in immune health. Join us on this transformative journey.
Research Projects
Determining Vaccine Responses Directly in Cancer Patients
Antibody titers to SARS-CoV-2 in infants (Wimmers, Cell, 2023).
Infectious diseases pose a significant threat to global health, with the top three alone accounting for over 12 million deaths in 2021. Cancer patients, particularly vulnerable, often succumb to infections and face diminished vaccine efficacy. The complexity and individual variability of immune responses in these patients remain largely uncharted territories. We aim to illuminate these hidden mechanisms, unraveling how cancer alters immune responses to vaccines.
For more information on how we use systems biology and mulit-omics tools to study immune responses, check out our work on immunity in cancer patients, and our studies to determine immunity to COVID and influenza vaccines and infections.
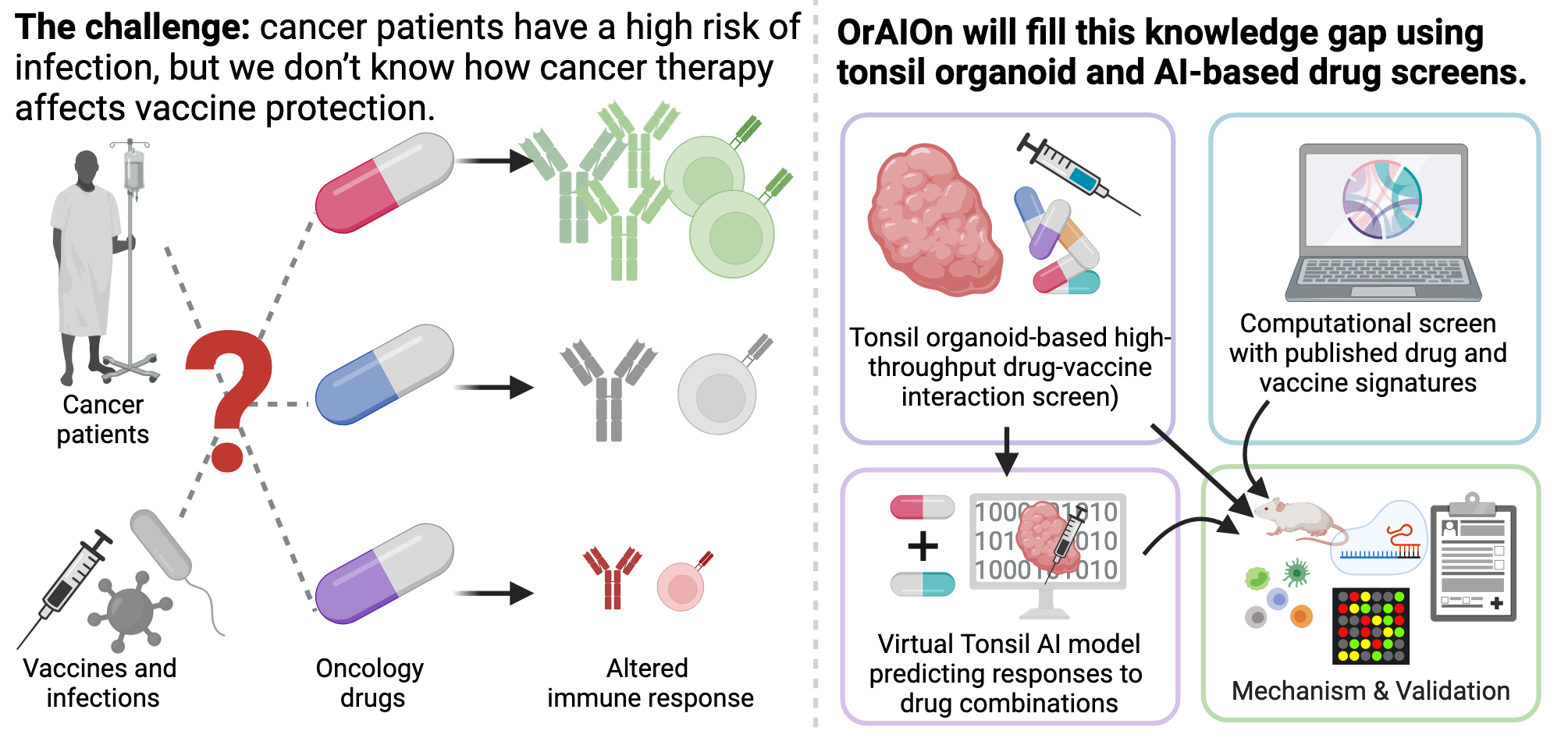
Understanding Drug-Vaccine Interactions in Cancer
Schematic overview of the ERC-funded OrAIOn project. (with biorender)
Florian on the OrAIOn project. Activate subtitles (German) for English translation.
IRecent studies suggest that some oncology drugs, while primarily targeting tumor progression, also influence immune functions, which can alter vaccine efficacy. Our new ERC-funded project, OrAIOn, aims to systematically investigate how a wide range of these drugs affect vaccination outcomes.
Using innovative approaches, including a human tonsil lymphoid organoid-based screen and computational analysis of extensive drug profiles, we will explore the interactions between oncology drugs and vaccines. Our work will result in the first comprehensive database of drug-vaccine interactions, providing mechanistic insights to guide future clinical trials, optimize vaccine strategies for cancer patients, and improve anti-cancer vaccine development.
Defining the Epigenomic Immune Cell Landscape
Vaccines imprint epigenomic and functional changes to immune cells. (Wimmers, Cell, 2021)
Recent studies hint at a fascinating possibility: vaccines might shield us not just from their targeted pathogens but a wide array of unrelated ones. Imagine a flu shot boosting defenses against Dengue and Zika. This intriguing phenomenon could be rooted in epigenomic changes within our immune cells. We aim to decode these vaccine- and disease-triggered epigenomic shifts and explore their impact on immune cell function. To achieve this, we use a range of bulk and single-cell techniques to study the genetic, epigenetic, and transcriptional circuits controlling these processes, including scATAC-seq, scRNA-seq, CUT&RUN, and EpiTOF.
For more information, check out our work on influenza vaccination (Wimmers, Cell, 2021), and the experimental vaccine adjuvant 3M-052 (Lee, Nat Comm, 2022), and our perspective on vaccines (Wimmers, Curr Opin Immunol, 2020).
Technologies we love
Multi-Omics Analysis
We envision that integrating multiple high-content datatypes within human specimens will unlock new insights into immune responses and lead to tailored vaccination strategies. We’ve previously used this approach to gain insights into immunity to infection and vaccination: Wimmers, Cell 2023; Arunachalam, Wimmers, Science, 2020.
Single-Cell Tools
We develop and use cutting-edge single-cell technologies to explore immune cell heterogeneity, a key factor influencing vaccine efficacy. By mapping diverse immune cell responses, we aim to predict and enhance vaccine effectiveness, ultimately paving the way for personalized immunization strategies. Wimmers, Nature Com, 2018; Wimmers, Cell 2021.
Immunological Tools
To assess the functional state of immune cells, we combine our genetic and omics data with a range of immunological techniques. These include high-dimensional cytometry, antibody-based protein detection assays like ELISA, and in-vitro models for cell activation and differentiation.
Complex Human Organoid Systems
We believe that advanced tissue and organoid systems hold the key to uncovering new mechanistic insights into human immunology and accelerating the design of personalized vaccines. To achieve this, we develop and utilize human organoids derived from lymphoid and tumor tissues to study interactions between the immune system, vaccination, and cancer.
High-Throughput Drug Screens
We use high-throughput drug screens to uncover unexpected interactions between drugs and the immune system. Combined with computational methods, this approach helps us rapidly identify drug effects on vaccine responses, guiding the development of personalized immunotherapy strategies.
Chromatin Analysis
We aim to decode epigenomic changes and their effects on immune cell function. To do this, we apply a variety of chromatin technologies, including scATAC-seq, scRNA-seq, CUT&TAG, and EpiTOF, to study the genetic, epigenetic, and transcriptional networks that regulate immunity. Wimmers, Cell, 2021; Lee, Scott, Wimmers, Nature Com, 2022.
What we achieved (so far):
of genetic sequencing data analyzed
immune cells collected from blood samples
16
students trained since 2023
~1,600
items of plasticware recycled in 2023
>20,000
cryotubes labeled
>20
papers published
seconds of work saved with Robin's program
39,000
6
nationalities in our team
>13 billion
>5.2TB
News & Publications
Reach out if you want to learn more about our work.
Systems Immunology Laboratory
Department for Molecular Medicine
Interfaculty Institute for Biochemistry
University of Tübingen
Auf der Morgenstelle 34
72074 Tübingen